DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology
Por um escritor misterioso
Descrição
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.
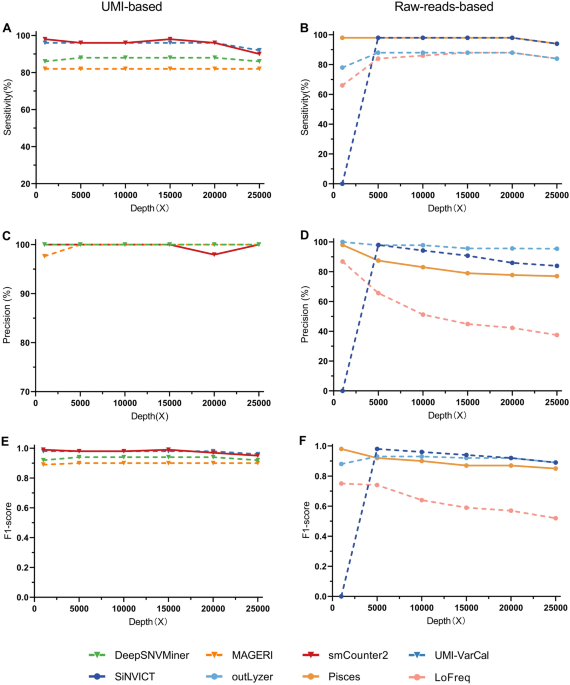
Evaluating the performance of low-frequency variant calling tools

Machine learning guided signal enrichment for ultrasensitive
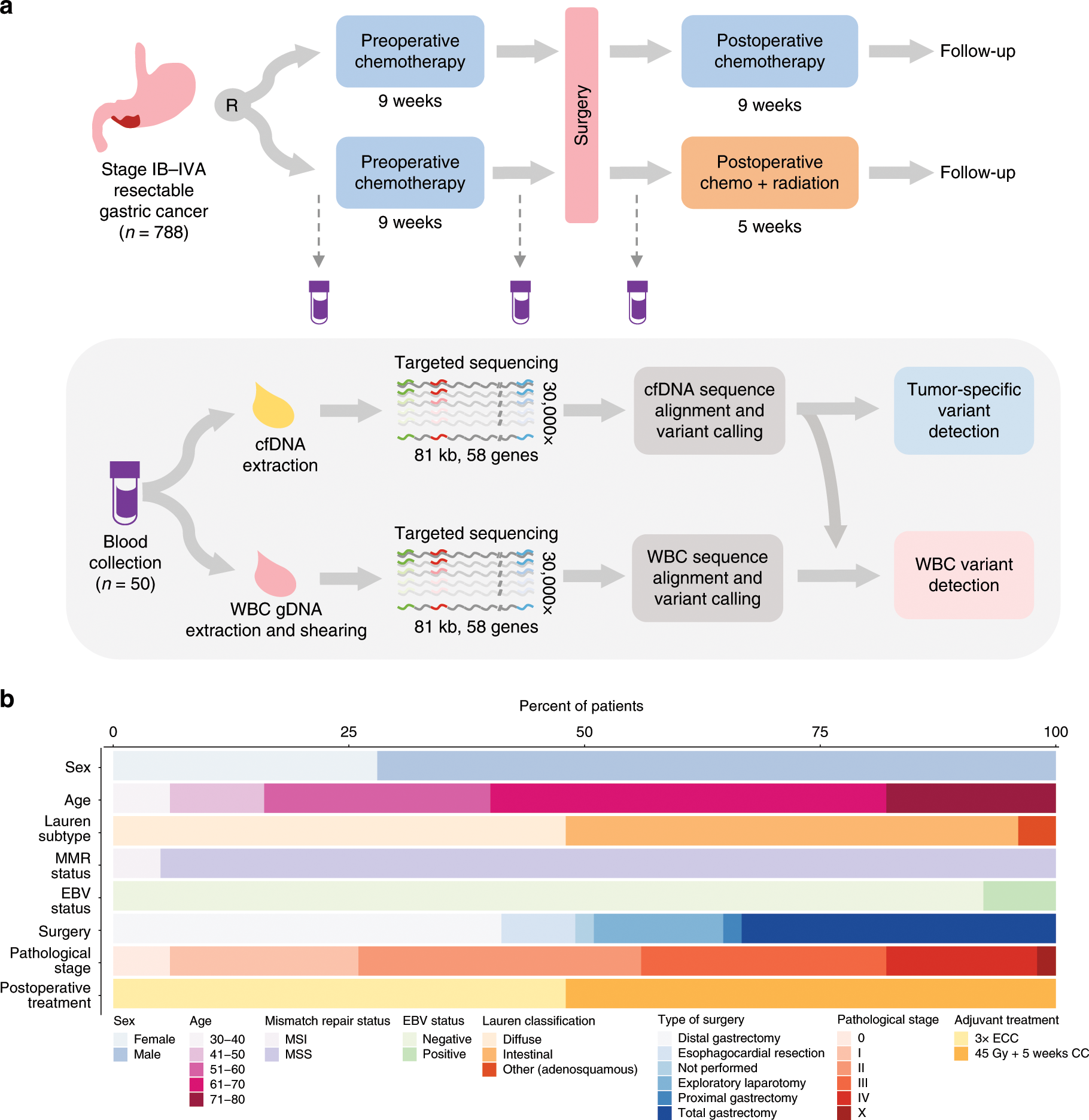
White blood cell and cell-free DNA analyses for detection of
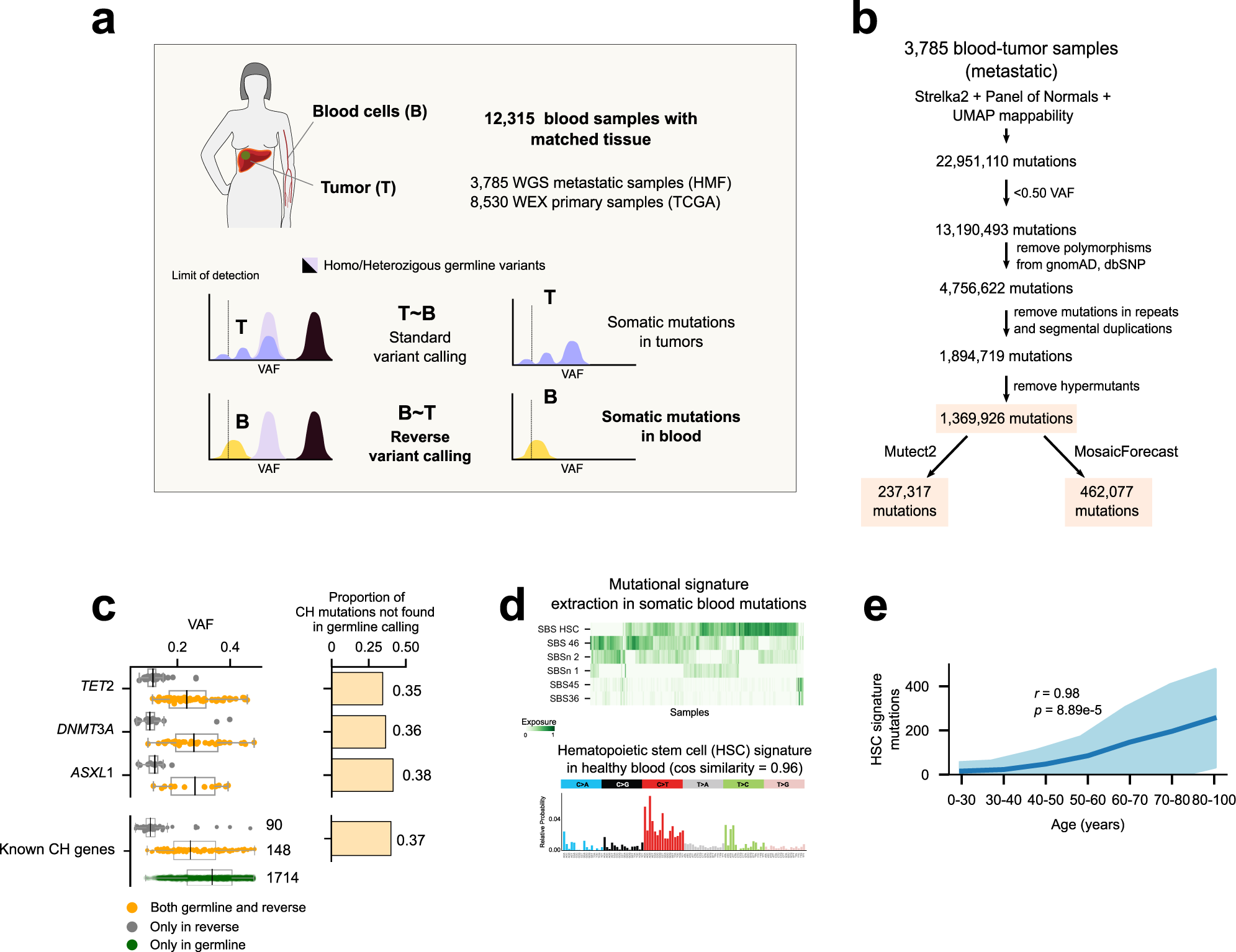
Discovering the drivers of clonal hematopoiesis
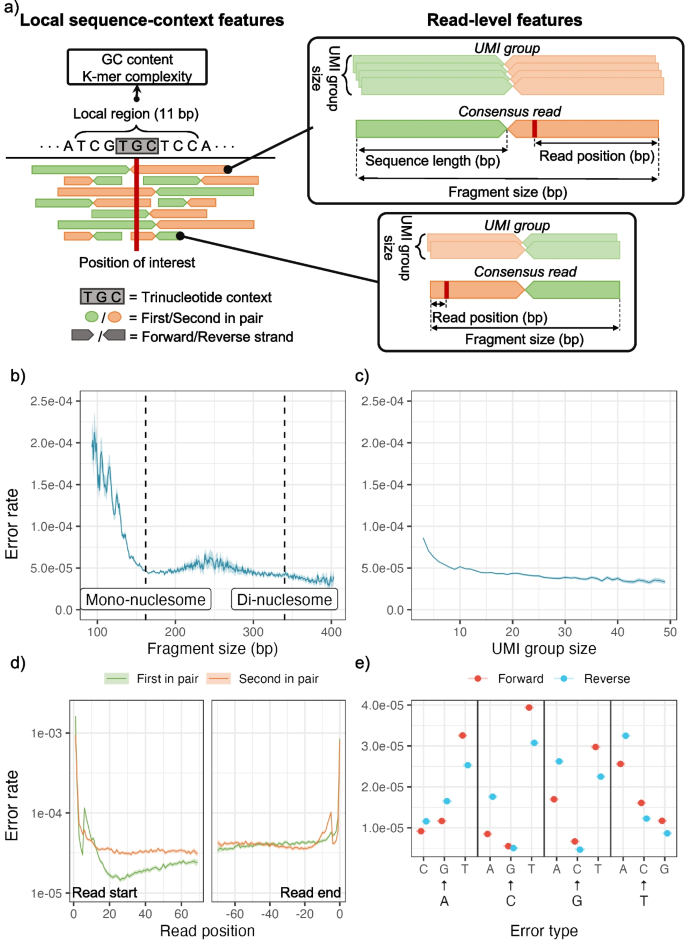
DREAMS: deep read-level error model for sequencing data applied to

Machine learning guided signal enrichment for ultrasensitive

PDF) DREAMS: Deep Read-level Error Model for Sequencing data
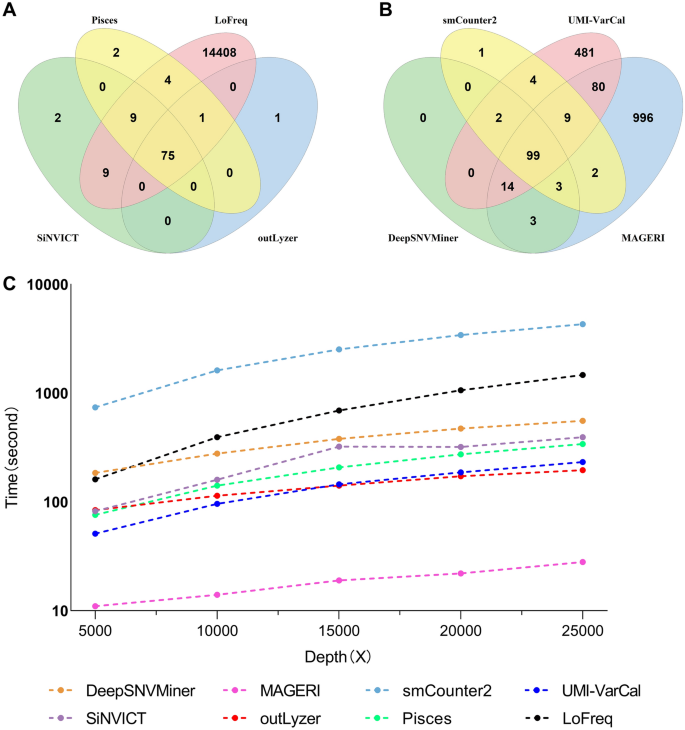
Evaluating the performance of low-frequency variant calling tools

LFMD: detecting low-frequency mutations in high-depth genome

Variant calling in control data. (a) Power (true-positive rate) of
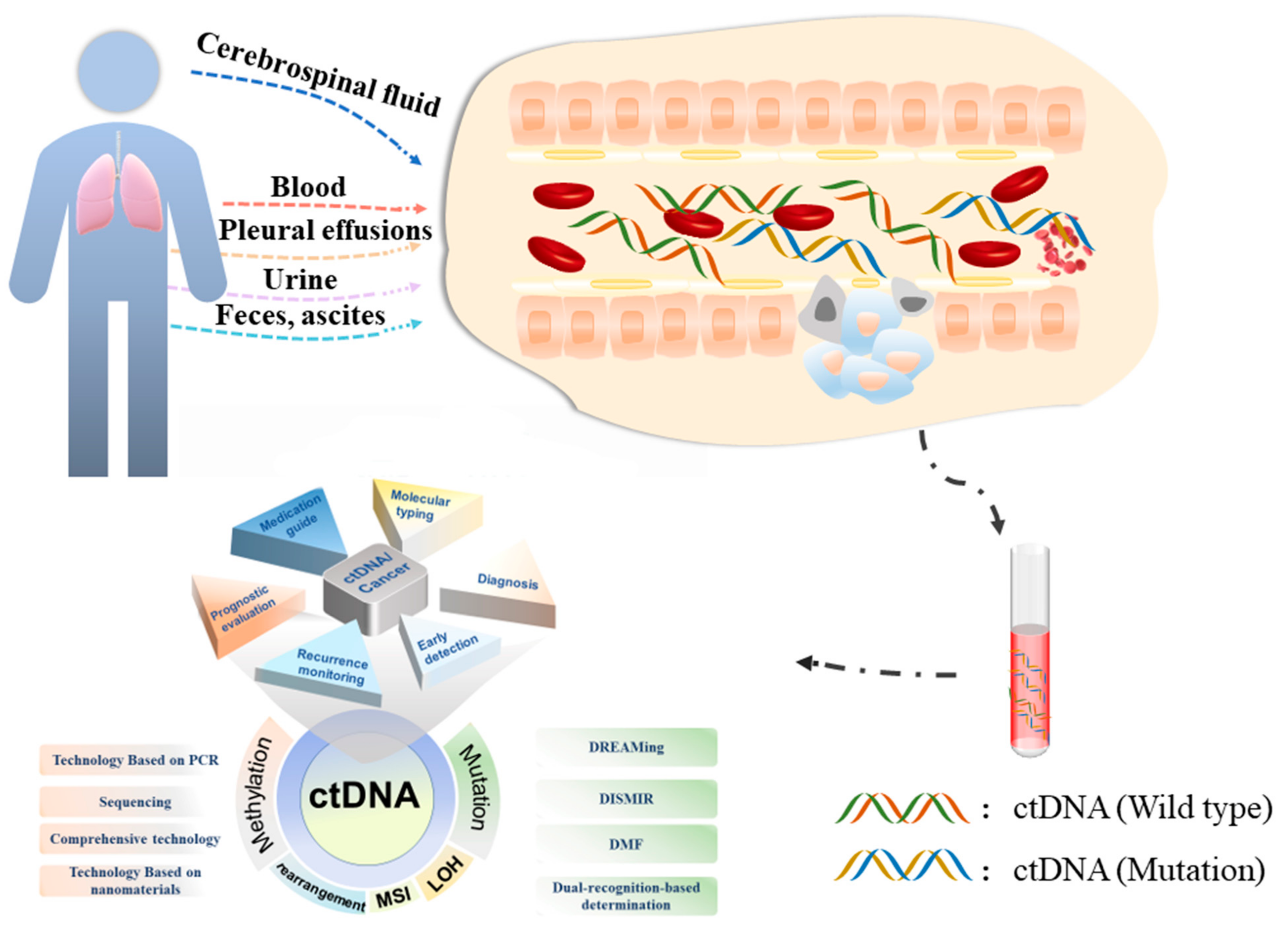
Cancers, Free Full-Text
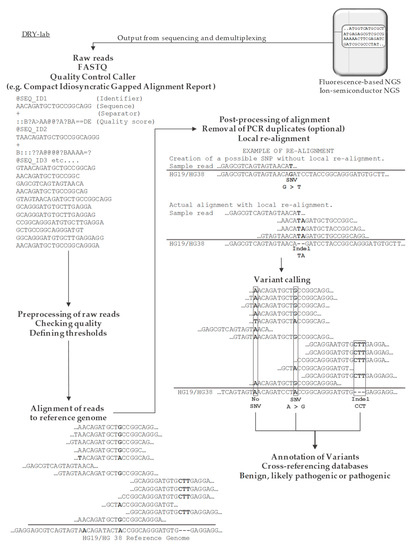
Cancers, Free Full-Text

Types of errors. A screenshot from the IGV browser [21] showing
de
por adulto (o preço varia de acordo com o tamanho do grupo)