Cells, Free Full-Text
Por um escritor misterioso
Descrição
Identifying tissue-specific molecular signatures of active regulatory elements is critical to understanding gene regulatory mechanisms. In this study, transcription start sites (TSS) and enhancers were identified using Cap analysis of gene expression (CAGE) across endometrial stromal cell (ESC) samples obtained from women with (n = 4) and without endometriosis (n = 4). ESC TSSs and enhancers were compared to those reported in other tissue and cell types in FANTOM5 and were integrated with RNA-seq and ATAC-seq data from the same samples for regulatory activity and network analyses. CAGE tag count differences between women with and without endometriosis were statistically tested and tags within close proximity to genetic variants associated with endometriosis risk were identified. Over 90% of tag clusters mapping to promoters were observed in cells and tissues in FANTOM5. However, some potential cell-type-specific promoters and enhancers were also observed. Regions of open chromatin identified using ATAC-seq provided further evidence of the active transcriptional regions identified by CAGE. Despite the small sample number, there was evidence of differences associated with endometriosis at 210 consensus clusters, including IGFBP5, CALD1 and OXTR. ESC TSSs were also located within loci associated with endometriosis risk from genome-wide association studies. This study provides novel evidence of transcriptional differences in endometrial stromal cells associated with endometriosis and provides a valuable cell-type specific resource of active TSSs and enhancers in endometrial stromal cells.
Cells, Free Full-Text

Challenging Post-translational Modifications in the Cell-free Protein Synthesis System - Synthetic Biology and Engineering - Full-Text HTML - SCIEPublish
Cells, Free Full-Text

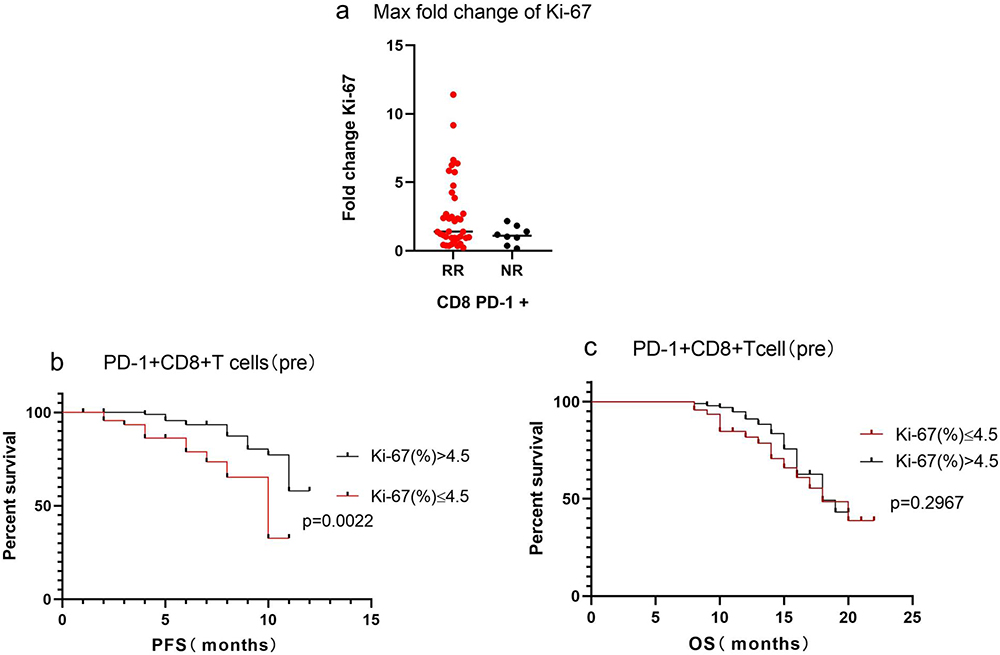
Cells, Free Full-Text
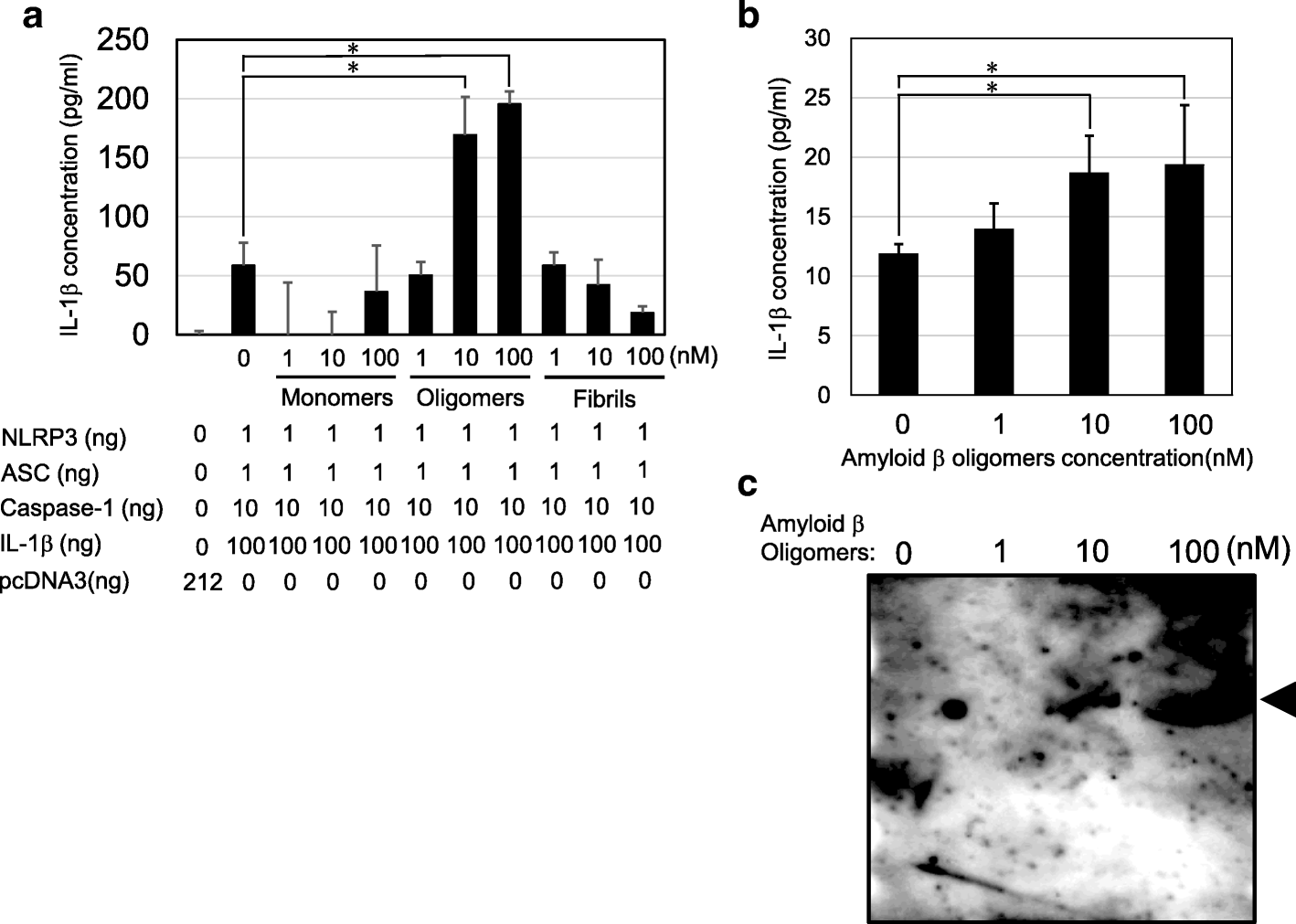
Amyloid β directly interacts with NLRP3 to initiate inflammasome activation: identification of an intrinsic NLRP3 ligand in a cell-free system, Inflammation and Regeneration
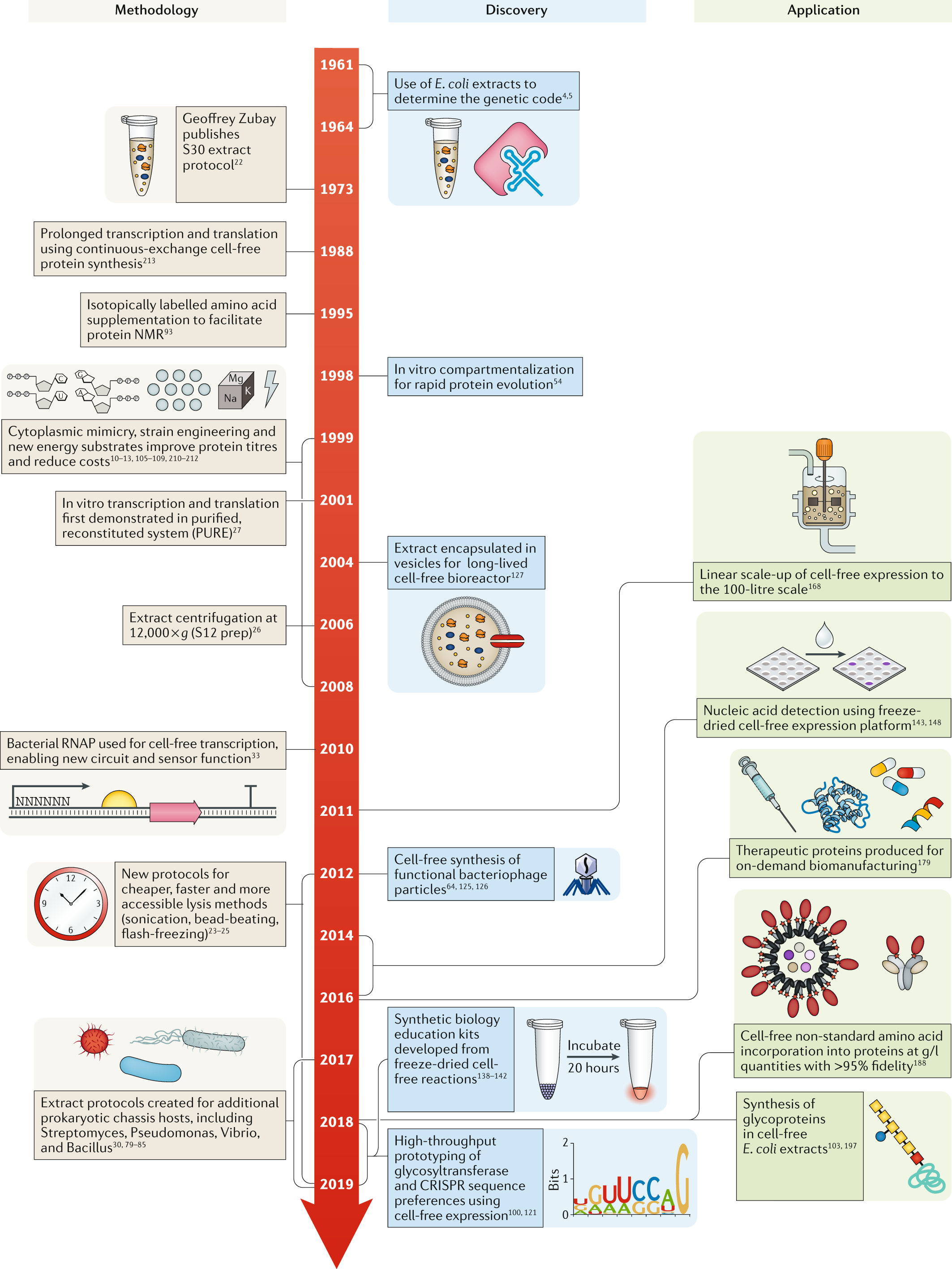
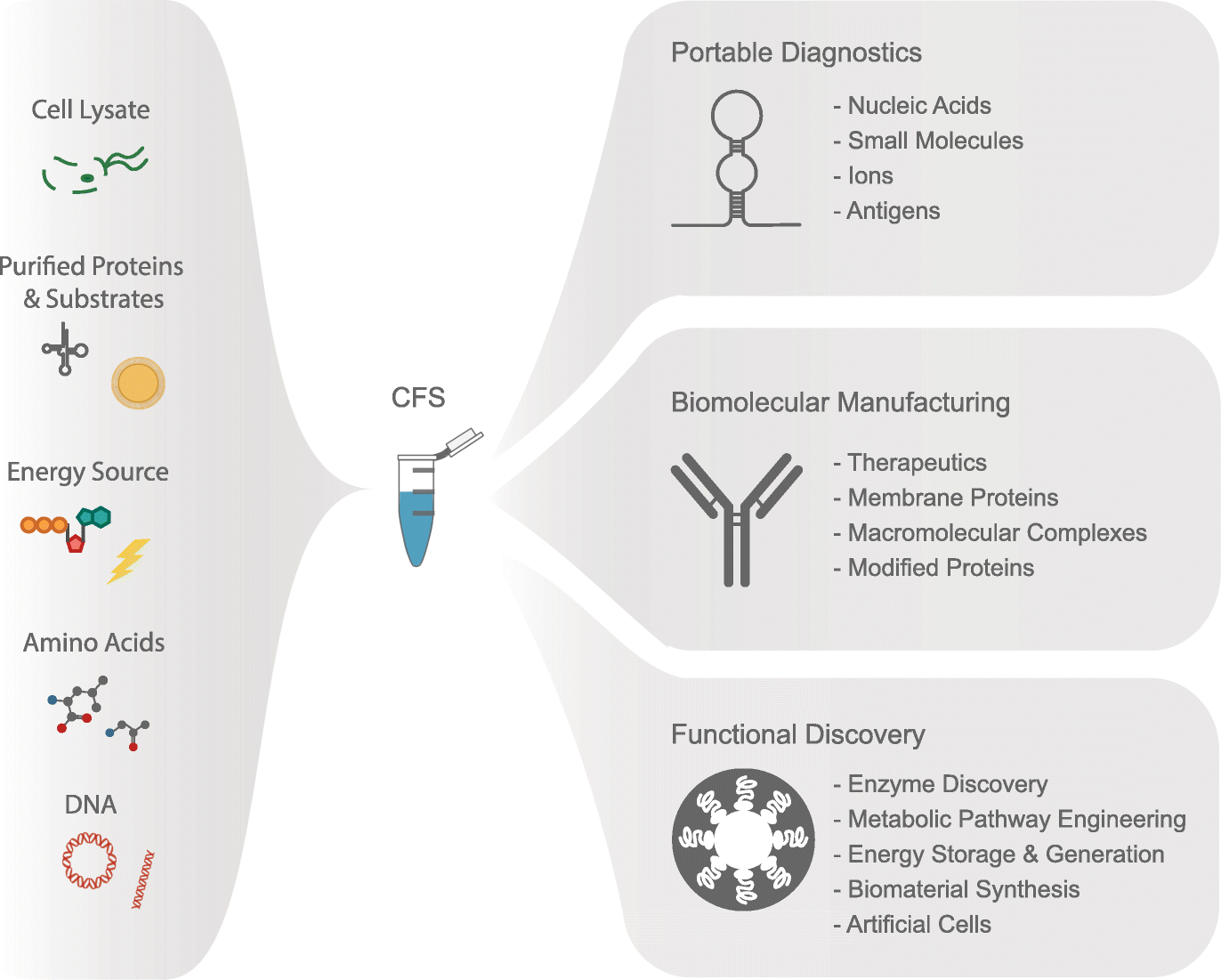
Cell-free gene expression: an expanded repertoire of applications

Harnessing Extracellular Vesicles for Regenerative Therapy - Gowing Life
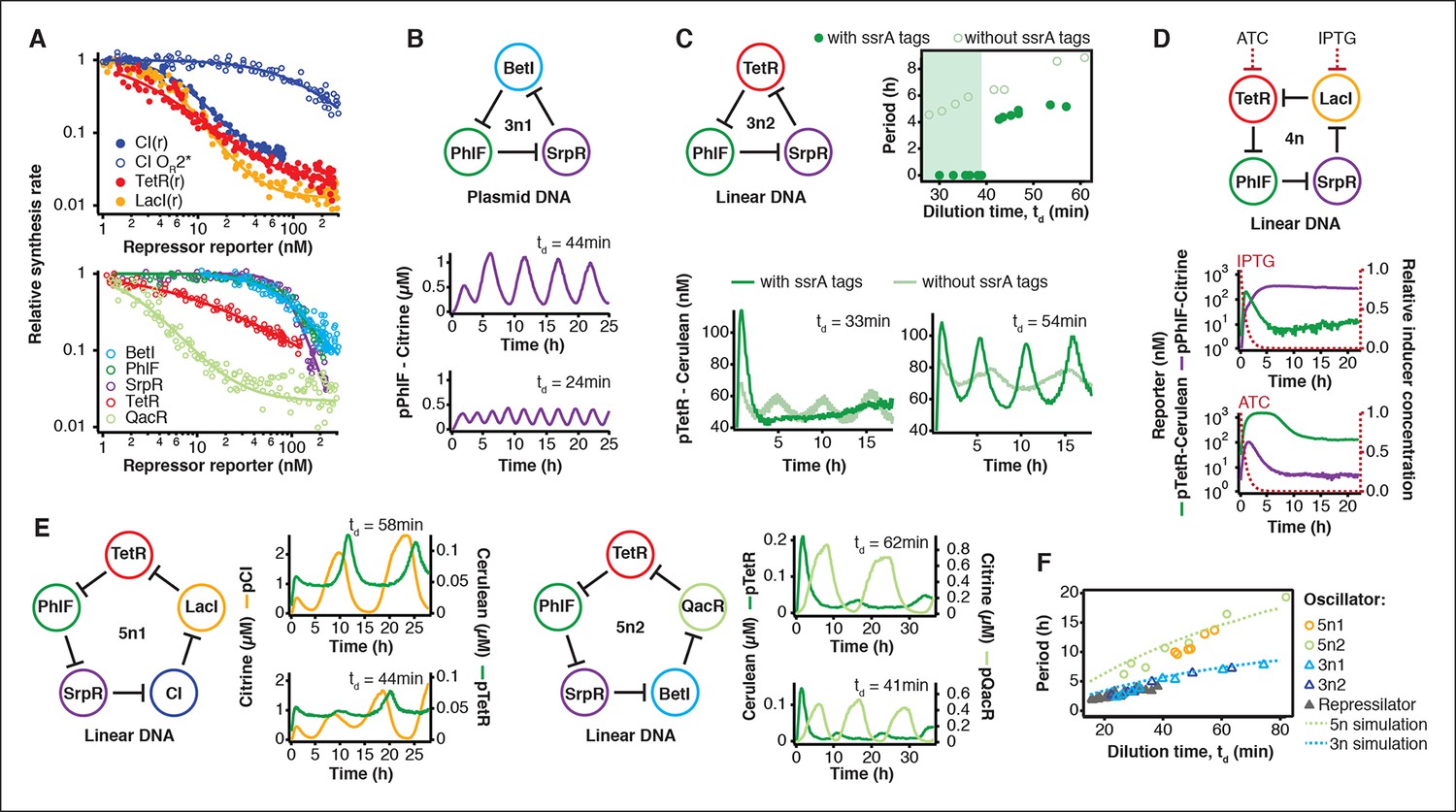
Rapid cell-free forward engineering of novel genetic ring oscillators

Sequencing of Circulating Cell-free DNA during Pregnancy
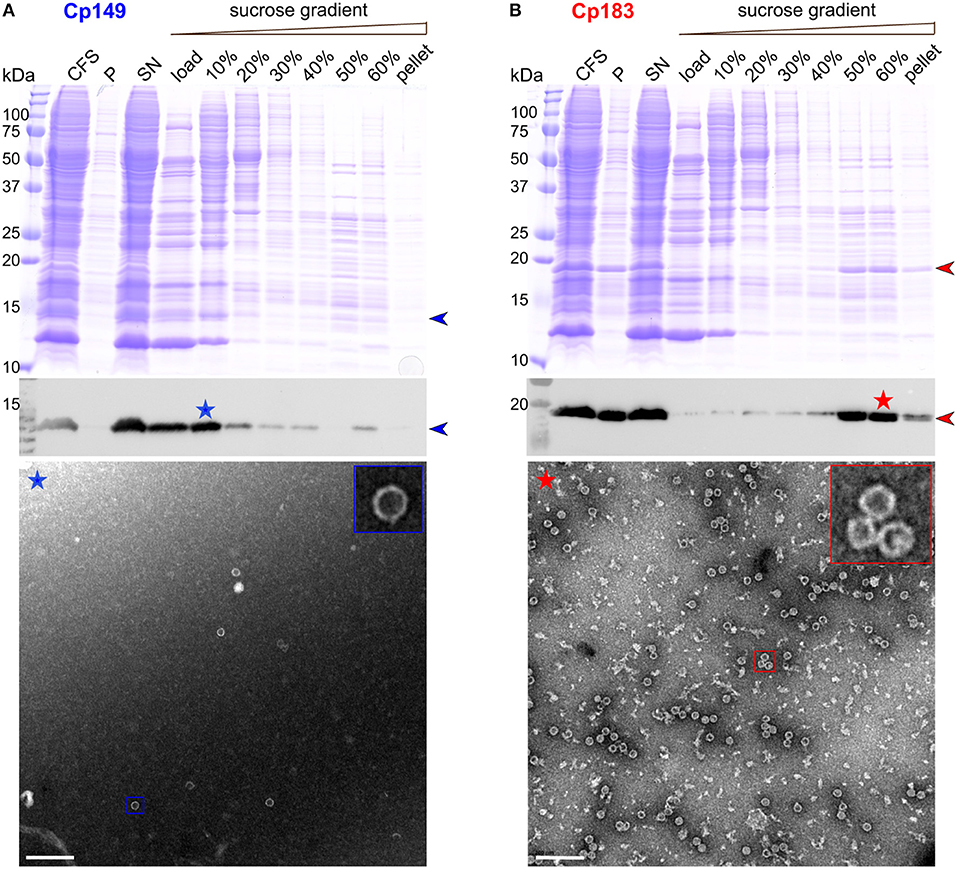
Frontiers Combining Cell-Free Protein Synthesis and NMR Into a Tool to Study Capsid Assembly Modulation
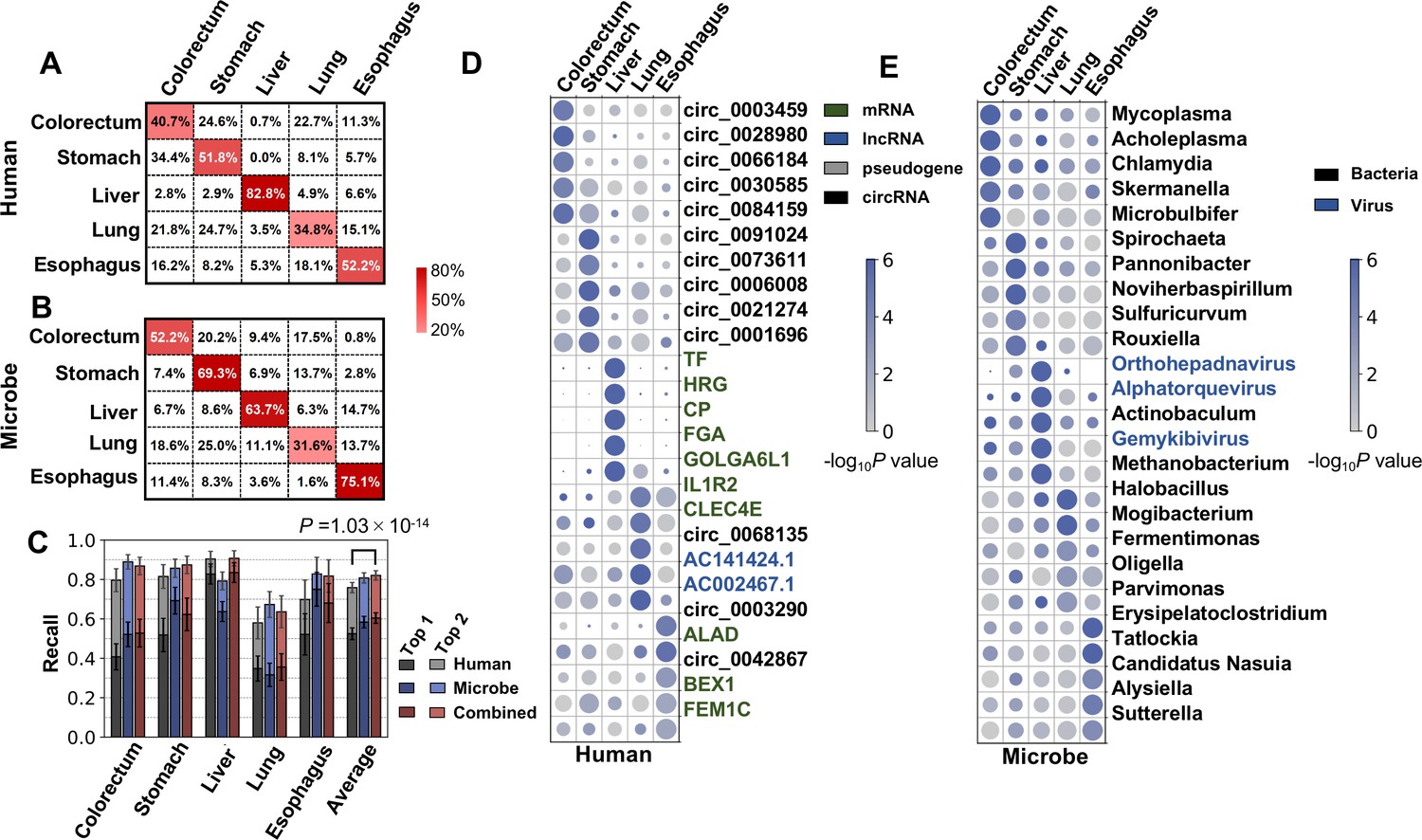
Cancer type classification using plasma cell-free RNAs derived from human and microbes
de
por adulto (o preço varia de acordo com o tamanho do grupo)